About This Project
Proteins do tasks crucial for life; hence defunct proteins cause disease, often originating from a mutation in an exon (DNA that becomes a protein). Identifying exons & mutations therein drive both diagnostics for identifying the best method of treatment & researchers by enabling them to elucidate the disease. Exons annotation is currently substantially sub-par; as a result most researchers ignore exons in analyses & there may be 10,000s more. I aim to identify them.
Ask the Scientists
Join The DiscussionWhat is the context of this research?
RNA sequencing (RNA-seq) - a form of genomic sequencing - has greatly dropped in price & its use has skyrocketed. It allows us to:
- use an individual's genome for personalized medicine, &
- identify genomic changes between conditions (e.g. healthy vs cancer)
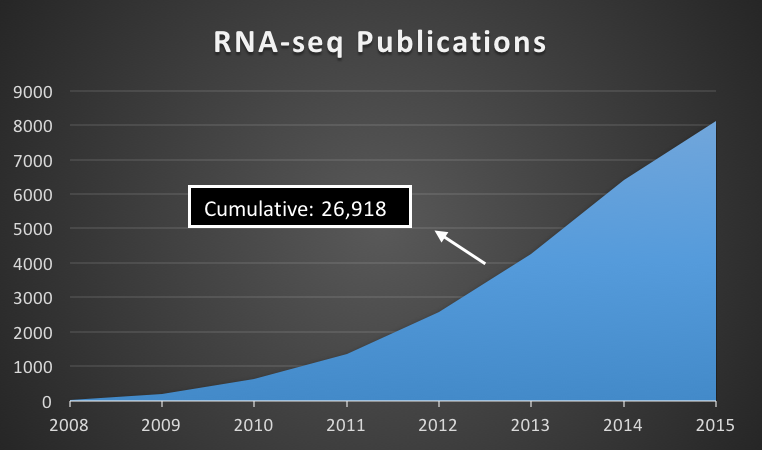
The former utilizes the knowledge built off the latter. The latter allows for both the detection of genes & exons (parts that make up genes) that change due to a condition (DEGs / DEEs respectively). However our annotation of exons is pretty bad... hence the ratio of DEE to DEG studies is about 0.23% (NCBI). Further, there may be 10000s more exons. This is unacceptable given that analysis of mutations in exons can allow us to expound a disease from its genetic origin.
Methods for plenty of details and graphics!
What is the significance of this project?
Why care about identifying and annotating exons?
Consider the clinical perspective - wanting better diagnostic capabilities. How can a medical practitioner identify a mutation in your genome if they do not know where to look? It is known that these variations may make certain medications ineffective for some individuals. This can prevent the prescription of less efficacious medicines.
Consider the scientific perspective, wanting to expound disease. How can scientists elaborate how mutations give rise to disease, if they do not know where those mutations occur and the relevance of that location?
Identifying exons allow for both scientists to better explain associated disease, and allow doctors to provide better and more curated medicine.
Methods for plenty of details and graphics!
What are the goals of the project?
While the technologies and methodologies implemented in the project are cutting-edge the logical progression of this project's implementation is not convoluted. Foremost, the machine on which the analyses will be run must be constructed. Then, the RNA-seq data must be gathered and preprocessed. Subsequently, features from the data can be extracted for use in the model.
Once we have the data squeaky clean, we can develop the deep neural network. Actually, we will build many, and then select the best one. To ensure our theoretical model is correct, we will confirm in vivo (in living animals). As this tool is meant to be used for others, we will lastly develop a web-applets for everyone to use.
Budget
Question: How did so much money get raised without any donors?
Answer: We had some off site donors :) Companies like NVIDIA, La Cie (now SeaGate), Deepcool, etc have donated hardware greatly reducing the gap to get this going
This research is deep-learning which requires a strong computational device. Thus it is self evident how these budget items go towards making this research possible by providing a device capable of the computations.
Endorsed by
Lab Notes
Nothing posted yet.
Additional Information
Project Backers
- 6Backers
- 78%Funded
- $11,641Total Donations
- $11.83Average Donation


