Methods
Summary
We will be performing In Situ Hybridization on penis tissue from developing mice, ferrets, and rabbits. Below is an outline of this process, which is also described in this video in plants.
Paraffin Embedding
- Tissues (genitalia) will be fixed in 10% Paraformaldehyde (PFA) to prevent degradation
- After fixation, genitalia will be dehydrated by soaking in increasing concentrations of ethanol
- Dehydrated tissues will then be placed in plastic tissue molds, and hot paraffin wax will be poured into the mold
- Once the wax is set in a block, we will carefully slice through the sample using a microtome. This will result in tissue sections between 5-10 microns thick
- Samples will them be mounted on glass slides
In Situ Hybridization
- We will amplify the Sox9 cDNA sequence through RT-PCR based on an RNA extraction from our mouse specimens
- This amplified sequence will then be inserted into a plasmid and expanded in yeast colonies
- After the further amplification of our target sequence, we will bind DIG-labeled probes
- The amplified Sox9 sequence bound to DIG will be hybridized to the developing mouse genitalia
- Antibodies that recognize DIG will then be bound to the target Sox9 RNA inside of the tissue
- The anti-DIG antibodies contain a key enzyme
- The tissue will then be exposed to a solution that activates the enzyme
- This enzymatic reaction results in turning tissue with our Sox9 RNA purple
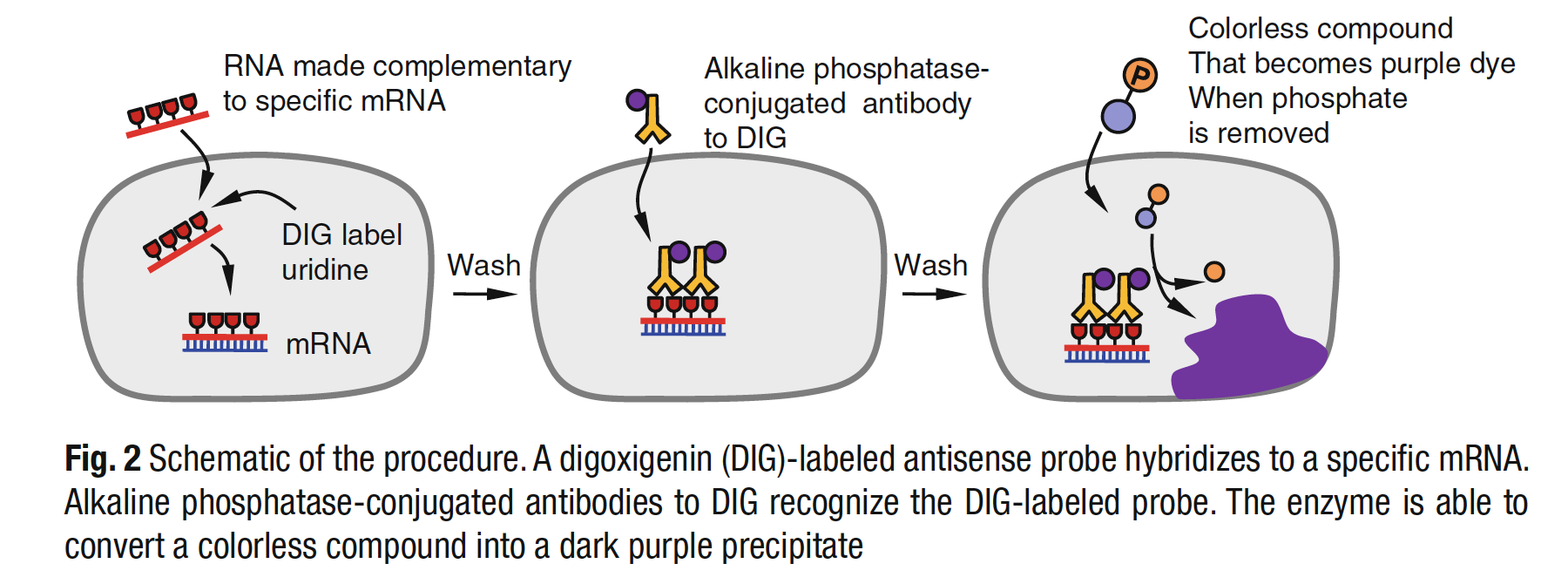 The above image is from: Lee, Daehoon, Shan Xiong, and Wen-Cheng Xiong. "General introduction to in situ hybridization protocol using nonradioactively labeled probes to detect mRNAs on tissue sections." Neural Development: Methods and Protocols (2013): 165-174.
The above image is from: Lee, Daehoon, Shan Xiong, and Wen-Cheng Xiong. "General introduction to in situ hybridization protocol using nonradioactively labeled probes to detect mRNAs on tissue sections." Neural Development: Methods and Protocols (2013): 165-174.
Protocols
This project has not yet shared any protocols.