About This Project
The high specificity and sensitivity of the CRISPR diagnostics, e.g. SHERLOCK, can detect the presence of the pathogen genome and report out the result accurately through the CRISPR-Cas system. It provides genome targeting based results and is superior to any serological based rapid test kits (RTKs). This project aims to improve the CRISPR diagnostic platform with an amplification-free approach, to enable a novel and robust RTK for SARS-COV2 detection.
Ask the Scientists
Join The DiscussionWhat is the context of this research?
Some Cas enzymes of the CRISPR system have collateral activity. This feature allows them to digest the nearby oligonucleotide after the CRISPR-Cas activation. Therefore, scientists apply this feature for pathogen identification.
There are two established CRISPR diagnostic platforms, namely Cas12-based DETECTR and Cas13-based SHERLOCK. Both of them required target amplification to increase the sensitivity of detection. Yet, this will extend the diagnostic duration.
Here we notice certain Cas enzymes can detect the target without amplification step. The identification of such Cas enzymes will ensure the development of an amplification-free CRISPR based rapid test kit.
What is the significance of this project?
The antigen-rapid test kit of SARS-COV2 can give false-negative results frequently. It also has lower sensitivity to detect the presence of SARS-COV2 compared to the PCR. Therefore, amplification-free CRISPR diagnostics can provide an alternative for fast and accurate SARS-COV2 rapid detection. We will know a passenger infection status in genome level within 15 minutes from the flight arrival to border checking.
We use a high sensitivity and novel Cas enzyme to develop an amplification-free fluorescent-illumination based rapid CRISPR diagnostic test kit for SARS-COV2 detection. It will push a leap in CRISPR diagnostics development, also provide the community with a fast, accurate and versatile test kit for viral detection.
What are the goals of the project?
Previously, we had identified the Cas enzyme that was able to detect the presence of a gene target with extreme sensitivity, together with its enzymatic kinetics profile.
Now we will evaluate the ratio of sgRNA, fluorophore-tagged oligonucleotide, buffer and Cas enzyme for the optimal reaction with the provided clinical samples and target oligonucleotide. Once the ratio of components is confirmed, we will freeze-dry the mixture to produce the lyophilised reactant for the test kit prototyping later.
The test kit prototype then will be assessed with a small scale of clinical samples. After that, we will bring the prototype for clinical trial later.
Budget
Previously we had established the proof of concept for the amplification-free CRISPR diagnostics via a self-funded budget. Here is the proposed fund allocation.
We need 600 dollars for the sgRNA synthesis and 400 dollars for the fluorophore-tagged reporter oligonucleotide synthesis. After the Cas enzyme reaction optimization of the sgRNA and fluorophore-tagged oligonucleotide has been done, we need 5000 dollars to construct the test kit prototype and validate it with a small pool of samples.
Endorsed by
 Project Timeline
Project Timeline
Once we receive the funding, we will engineer and construct a palm-warmth up reaction tube. The test kit prototype will be assembled for the trial with the collected clinical samples before early February 2022.
We will reoptimize the test kit accordingly, then validate the test kit through clinical trial according to the hospital protocol and regulations later by the end of February 2022.
We will manufacture this test kit massively and use it for clinical routines by March 2022.
Dec 14, 2021
Project Launched
Jan 01, 2022
Expected fund incoming
Jan 15, 2022
Clinical trial application submission
Jan 20, 2022
Palm-warmth up reaction tube engineering and construction
Feb 04, 2022
Trial with the collected clinical samples
Meet the Team
Ang Kuan Ping
Currently, I am a medical microbiology trainee at the University of Malaya.
Major interests
CRISPR biology: I focus on Cas enzyme mining through the bioinformatics approaches. Hopefully, there will be a novel Cas enzyme to be discovered. It will grant us new knowledge to understand the bacterial genome and physiology. It also provides us with a new tool for biotechnology application development, e.g. CRISPR diagnostics.
Developmental immunology: I work on the roles of RUNX1 in lymphocyte development. At the same time, I am interested in memory lymphocyte development.
Kc Liew
I have finished Infectious Diseases (ID) training from NSW. Currently, I am a Microbiology Registrar in Royal Melbourne Hospital.
I am one of the principal investigators involved in the Immunofluorescence serological assay for Whipple Disease. That project has collaborated with the Australian Rickettsial Reference Laboratory, Barwon Heath.
Currently, I am collaborating with the University of Malaya to develop CRISPR as a SARS-CoV 2 clinical diagnostic tool, improving the NGS bioinformatics pipeline. I am working on the functional assay to evaluate a gene in Pseudomonas aeruginosa that may be associated with higher mortality in patients with Quadram Institute Bioscience.
Jamal Sam
I am a clinical microbiologist in a busy teaching hospital in Kuala Lumpur, Malaysia. I am head of the diagnostic virology unit, which provides critical diagnostic support for COVID-19 patients, including rapid antigen detection, PCR, and whole genome sequencing. COVID-19 diagnostics incurs a considerable cost burden to resource-limited settings such as ours, and it is imperative to develop new diagnostic tools that are accurate, affordable and easy to use.
Won Fen Wong
Our research focuses on elucidating mechanism of immune system against different pathogen infections. Following COVID19 pandemic, our team is exploring a potential usage of CRISPR technology mediated RNA cleavage for viral detection and elimination purposes.
Additional Information
What had we done so far?
This project started in January 2021, when we noticed the possibility of amplification-free CRISPR diagnostics with certain Cas enzymes. We spent six months (January 2021 - June 2021) evaluating the usage of different synthetic oligonucleotide sequences as targets and the compatible sgRNAs to obtain the enzymatic kinetics profile. We completed the proof of concept stage for the amplification-free CRISPR diagnostics.
After that, we developed the platform foundation for SARS-COV2 detection. We identified the target sequence from the SARS-COV2 genome, then synthesized the corresponding sgRNA. We validated the sgRNA with the synthetic target sequence and three clinical samples for the preliminary result.
The preliminary result shows that our Cas enzyme candidate can detect the presence of the target effectively in 15 minutes without the amplification step. Consequently, we are confident to apply our Cas enzyme candidate for the target detection without the amplification step.
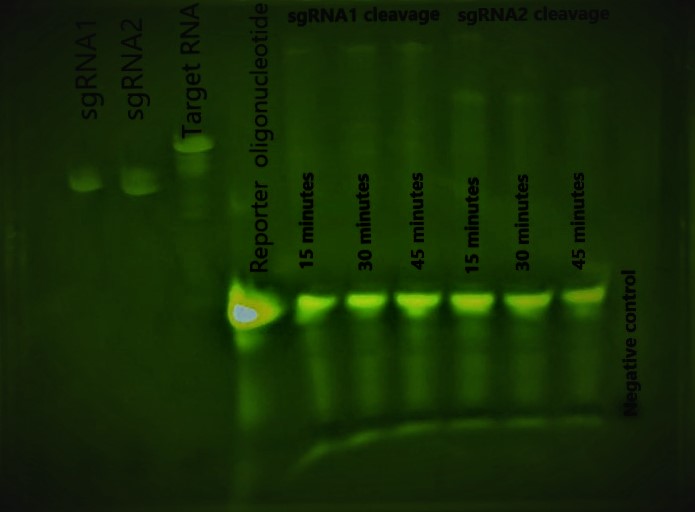
In addition, we conclude from the preliminary result that our Cas enzyme candidate can detect the presence of the target in picomolar concentration. The reported result is comparable to the standard PCR result with high accuracy.
.jpg)
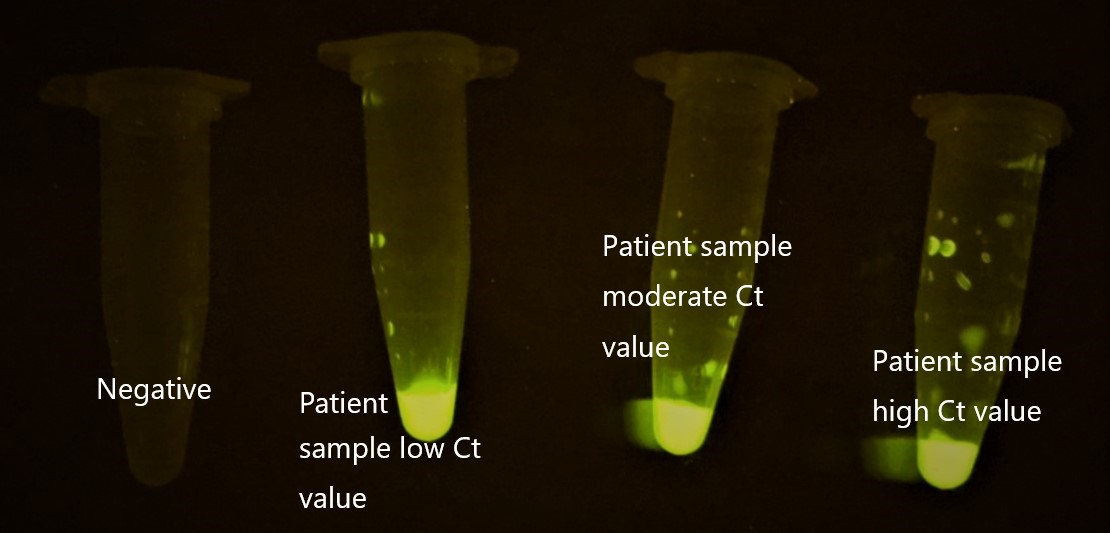
We tried to detect the SARS-COV2 from the patient samples with our CRISPR diagnostic platform. Following is the result. Negative remained while the samples with different Ct values, all detected.
Project Backers
- 19Backers
- 100%Funded
- $6,040Total Donations
- $317.89Average Donation




