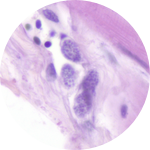
About This Project
Due to their cryptic nature, very little is known about the northward-creeping and vector-borne pathogens that cause malaria (Plasmodium odocoilei), babesiosis (Babesia odocoilei), or theileriosis (Theileria cervi) in cervids. These pathogens are moving north along with their mosquito and tick vectors and white-tailed deer hosts due to climate and land use changes. These pathogens pose a threat to northerly-distributed and declining moose, reindeer, and caribou populations.
Ask the Scientists
Join The DiscussionWhat is the context of this research?
White-tailed deer are expanding their geographic range northward and increasing in abundance with changes in climate and human land use. White-tailed deer are competent reservoir hosts for a diversity of pathogens vectored by mosquitoes and ticks which are also increasing in range and abundance. As white-tailed deer, mosquitoes, and ticks expand northward and become more common, they increasingly overlap in range with northerly-distributed and declining cervid species vulnerable to pathogen spillover and subsequent disease emergence including moose, elk, and reindeer. Naïve hosts such as elk and reindeer often die from B. odocoilei or T. cervi infections, and we are still learning about the health effects of the recently re-discovered P. odocoilei.
What is the significance of this project?
Without comprehensive pathogen surveillance, we miss the opportunity to take a proactive management approach rather than a reactive approach to pathogen spillover and subsequent emerging infectious disease outbreaks in wildlife. Establishing background information on P. odocoilei, B. odocoilei, and T. cervi will allow us to evaluate spillover risk to northerly-distributed and immunology-naïve ungulates such as moose and reindeer as the pathogens continue to move northward. We hope the results of this study will inform the decisions of wildlife managers, biologists, veterinarians, and livestock owners and aid in preventing the spillover of dangerous pathogens to populations of conservation concern. The project may also elucidate concerns for the health of livestock and humans.
What are the goals of the project?
The primary goal of this project is to establish baseline data on the geographic and host distribution and prevalence of P. odocoilei, B. odocoilei, and T. cervi in cervids throughout the northeastern US. This is a crucial step in understanding how these pathogens are shifting across time, latitude, and species in response to climate change. By PCR analysis, we will screen samples obtained from free-ranging moose and white-tailed deer from Connecticut, Massachusetts, Vermont, New Hampshire, Maine, and New York. We will also screen samples from captive cervids in the same states, including white-tailed deer, reindeer, elk, red deer, fallow deer, and other species. Hundreds of samples have already been procured from our collaborators.
Budget
Through broad collaboration, we have obtained hundreds of cervid tissue samples along a broad latitudinal gradient (Connecticut to Maine) ranging in collection date from 2007 to 2022. The funds from this campaign will allow for PCR screening of these samples for the pathogens P. odocoilei, B. odocoilei, and T. cervi. This study will generate critical background information on these cryptic and northward-moving vector-borne pathogens. Results will elucidate how these pathogens are distributed geographically and how their ranges have changed over time.
Endorsed by
 Project Timeline
Project Timeline
This project will run for approximately a year and a half.
Jun 01, 2022
Project Launched
Aug 01, 2022
Continue to receive and organize archived samples from collaborators
Dec 01, 2022
Opportunistically collect samples from necropsy, roadkill, and hunting check stations
Jan 01, 2023
Screen samples using molecular methods for parasite infection
Mar 01, 2023
Sequence DNA to identify parasites
Meet the Team
Affiliates
Team Bio
We are part of a group of researchers comprised of faculty, graduate students, and undergraduate students in the Wildlife Pathogens Laboratory at the University of Vermont. We investigate wildlife pathogen spillover, discovery, and systematics. You can read more about our work here.
Lauren Berkley
Lauren is currently pursuing her M.S. in Biology at the University of Vermont, where she graduated with her B.S. in Wildlife Biology in 2020. She has a wide variety of experience in working with wildlife, ranging from wolf education and care to marine mammal rescue and rehabilitation to protected shorebird research and management. She is broadly interested in wildlife disease ecology. More specifically, she is focused on the spillover of wildlife diseases to novel wildlife hosts, livestock, and humans with the progression of climate change and habitat degradation. She enjoys working with diverse collaborators to develop One Health solutions to our most pressing health challenges.
Ellen Martinsen
Dr. Ellen Martinsen is a biologist with research interests in host-parasite ecology and evolution, wildlife disease, and conservation. She is an Adjunct Assistant Professor in the Department of Biology at UVM and a Research Associate at the Smithsonian Conservation Biology Institute’s Center for Conservation Genomics. She has investigated the diversity and distribution of malaria parasites in various bird, mammal, and mosquito (vector) communities worldwide. Along the way she has discovered numerous new parasite species and host-parasite associations, endemic New World mammalian malaria parasites, and lots of host switching. Through broad collaboration and the integration of field, microscopy, and genetic methods, she has pursued the study of pathogen spillover into naive host taxa including collection animals at zoological parks, host species of conservation concern, and non-adapted wildlife species at northern latitudes.
Project Backers
- 74Backers
- 119%Funded
- $3,148Total Donations
- $29.03Average Donation