Methods
Summary
OVERVIEW
Part of the beauty of this project is that it uses terabytes of previously published data and processes it together to have massive impact at a small fiscal cost. The low cost stems from the fact that the majority of this project will be done in silico (done in / on the computer). Thus the techniques are all pretty straightforward. In short, an without trying to explain probabilistic models without a LaTeX extension, I will be applying a bayesian neural network.
For those unfamiliar with neural networks, information theory, inference, and bayesian networks feel free to consult one of the many available books on the subject e.g. Bayesian Neural Networks, Neural Networks and Learning Machines, Deep Belief Nets in C++ and CUDA C, or Information Theory, Inference and Learning Algorithms.
For those who want to see recent applicant of these techniques, check out the work by the Frey Lab in Toronto. They have a series of high level reviews and publications in Bioinformatics and NIPS.
- A review of machine learning in genomic medicine.
- An example of deep learning performing well with genomic data Deep learning of the tissue-regulated splicing code
ARTIFICIAL NERUONS / NEURAL NETWORKS
Below is a stylized image of neuron. I'm highlighting four main parts:
- Soma
- Dendrites
- Synaptic Terminals / Buttons
- Axon
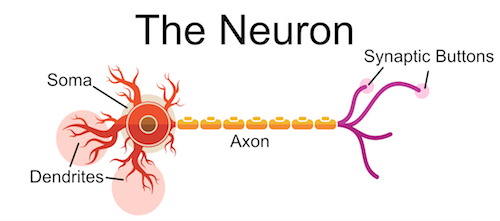
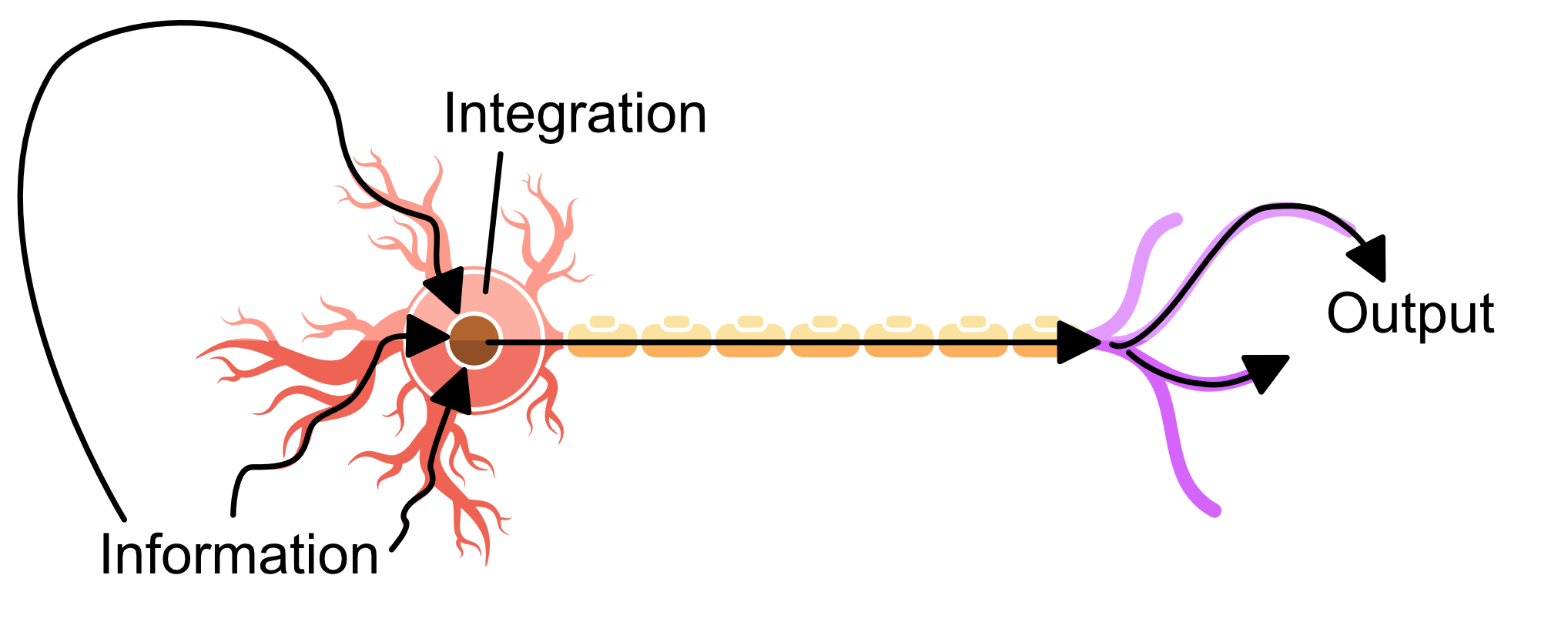
One could think of this neuron as an antenna, gathering information from the dendrites, integrating it at the soma, sending in it to a destination via the axon and finally transferred at the synapse. Below I highlight how the information flow's through the neuron.
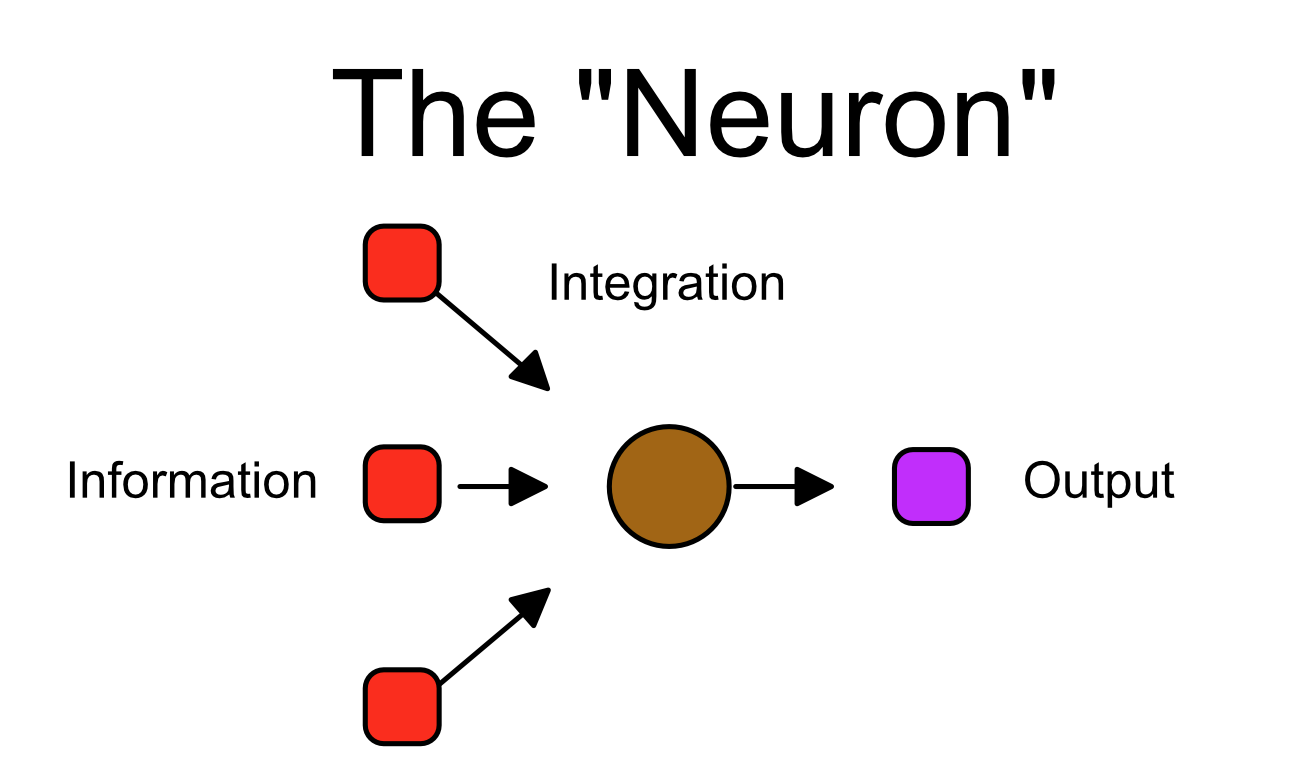
Using the main colors as in the previous images, we can abstract our neuron as such:
Then we can sprinkle on a little bit of math:
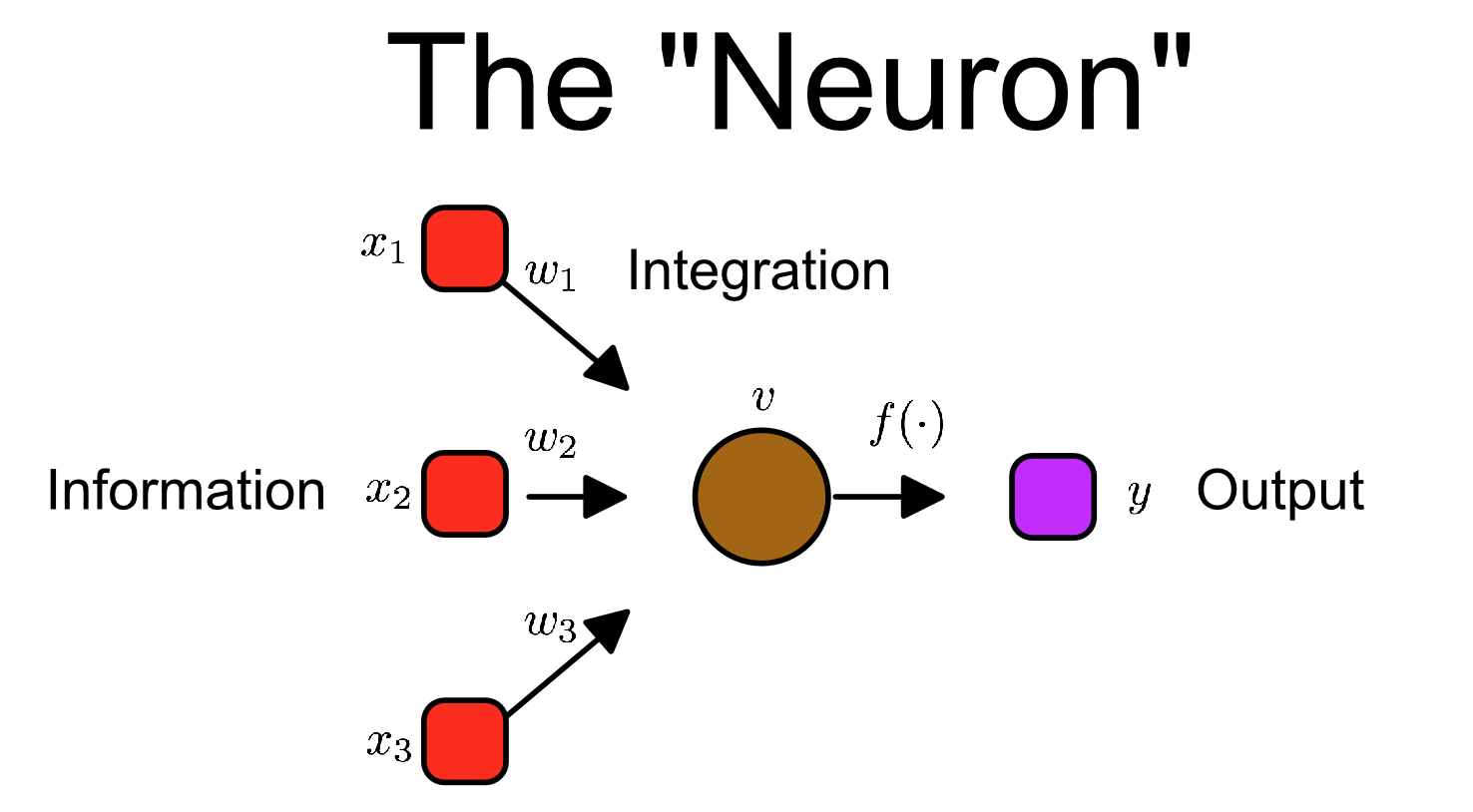
This concept of information flow through a neuron can be summarized in the following notation:
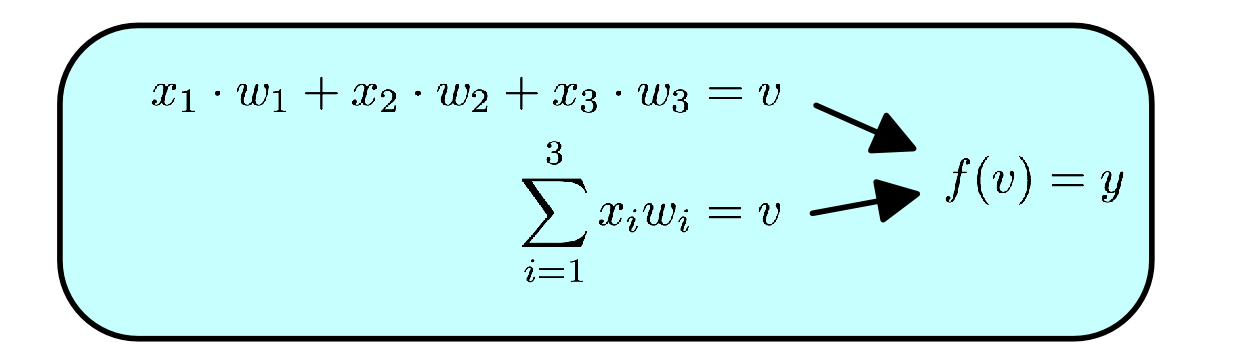
or in vector notation:
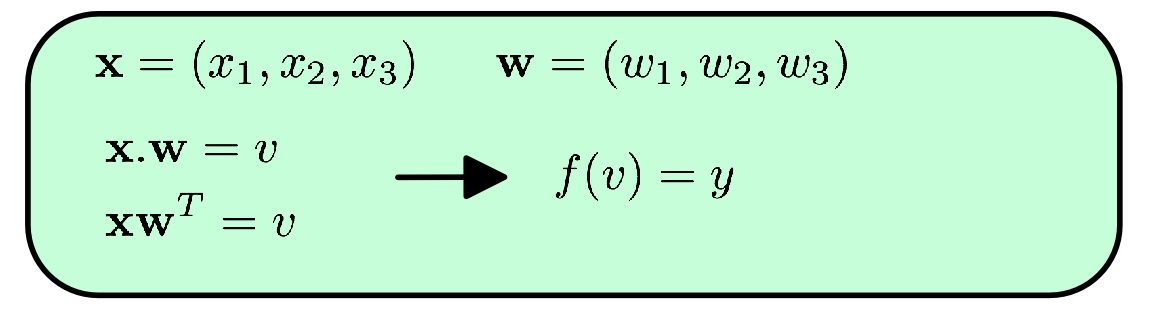
Not too scary! A (theoretical) neuron is essentially the sum of its inputs times their respective weights. Yes there are some nuances (like the squashing-function f which limits the neurons output. It just makes it easier computation), but really that right there, X.W - that simple equation - is the basis of neural networks.
Just as we abstracted a biological neuron to get an artificial one, we can do the same for a neural network. Where a biological neural network is a collection of neuron sending/receiving signals to/from one another, an artificial neural network (ANN) contains many artificial neurons (as described above) where the outputs of some neurons are the inputs to others.
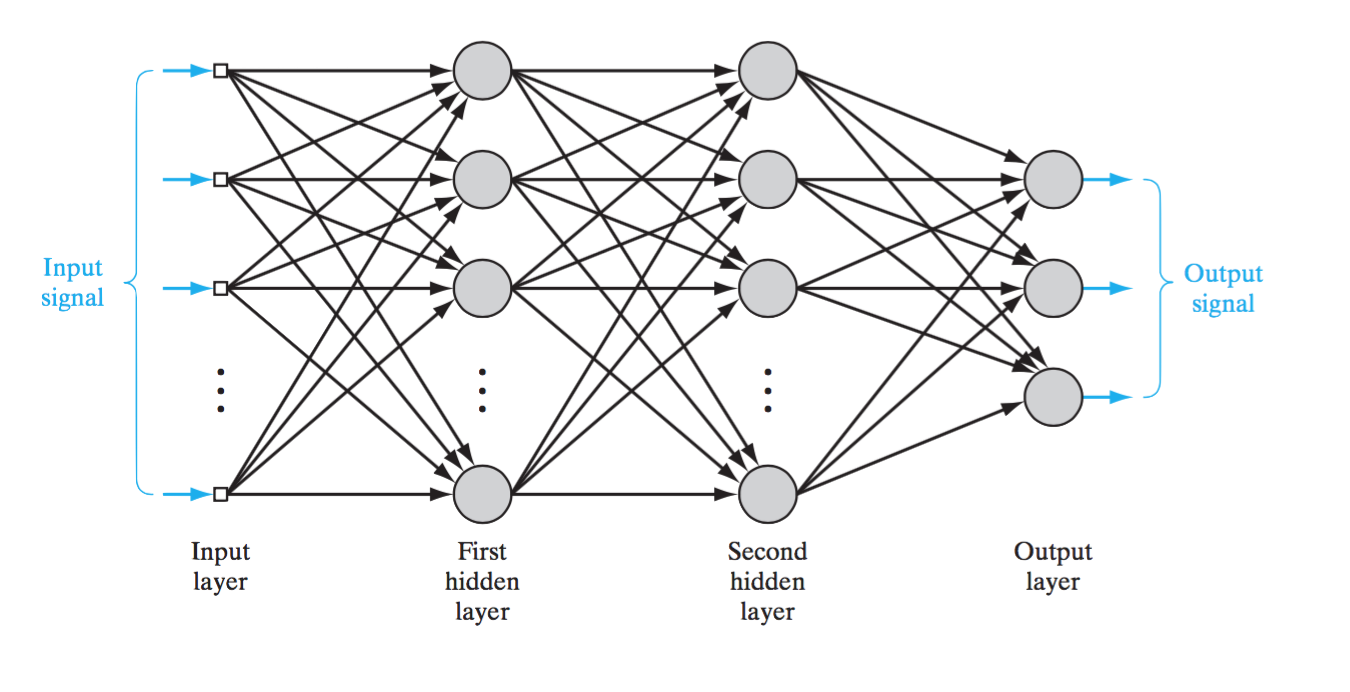 This remarkably reflects some aspects of the brain very well. Foremost is feature abstraction. As we go from the input layer to the output layer we sequentially abstract features of the data. There are other ways to neural networks can abstract features. For example, below is a convolutional neural network. Here we take our input layer and restructure it into a grid.
This remarkably reflects some aspects of the brain very well. Foremost is feature abstraction. As we go from the input layer to the output layer we sequentially abstract features of the data. There are other ways to neural networks can abstract features. For example, below is a convolutional neural network. Here we take our input layer and restructure it into a grid.
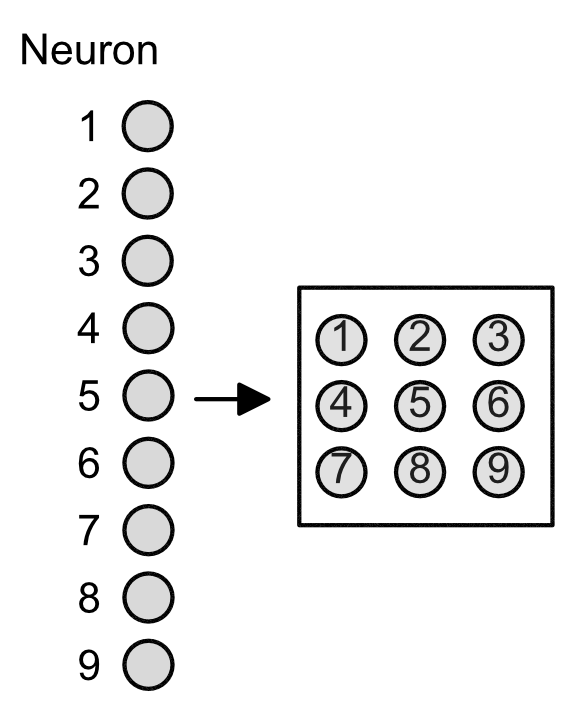
This grid allows for there to be local receptive fields (LRFs), illustrated below in the various colors. The next layer (hidden layer) is a single neuron representative of the field.
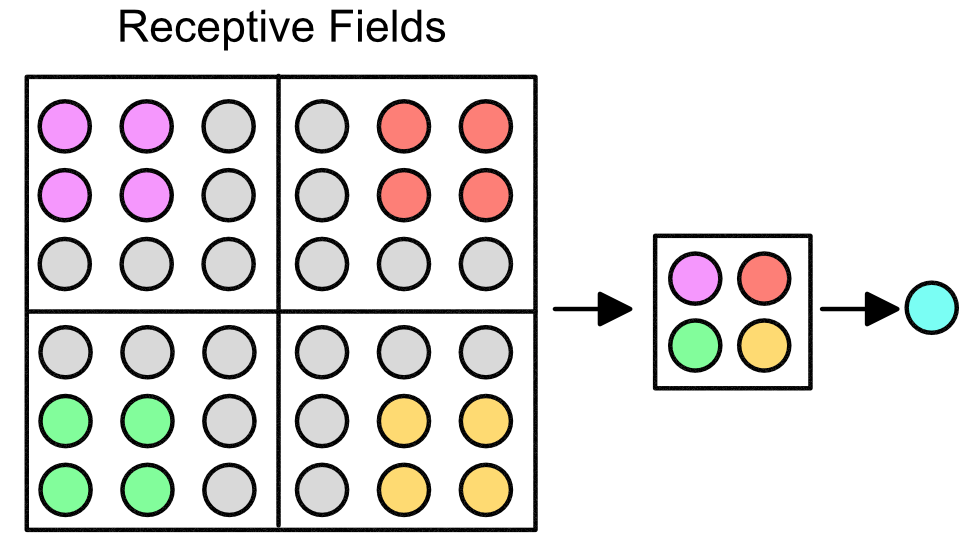
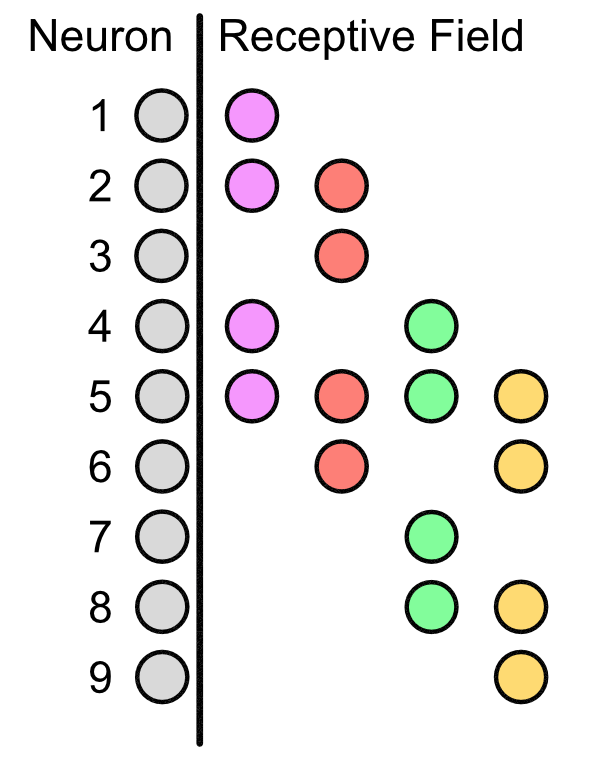
The importance of LRFs is that they allow for "scanning" of a sequence. Suppose that our original 9 input neurons represented features in our DNA. Then the LRFs allows for the hidden layer to get information across the entire sequence, e.g. see how the the neurons in the purple LRF correspond to neuron 1, 2, 4, and 5.
 A clear advantage of this is that the LRF can get more abstract features. For example, let's look at a picture of my family's dog, Ellie May.
A clear advantage of this is that the LRF can get more abstract features. For example, let's look at a picture of my family's dog, Ellie May.

While we process this image as a whole, as data it is stored as a sequence of bits (kind of like the DNA of the picture), going left to right, then starting a new row. The parts in the picture that define Ellie's ears are not next to linearly each other. So a linear network may not capture such a feature immediately, but LRFs can capture this feature as it is not looking for linear sequences.
If you're a bit lost. That is ok. Consider how DNA has features like promoter binding sites upstream of a TATA box. We know that some transcription factors or promoters can bind quite far away because of the 3D structure of DNA bending to allow contact. Thus it makes sense to not look just at the linear sequence.
Another key aspect of LRFs is that they share weights i.e. every neuron in that small 2 x 2 box has the same weight. Why might this matter? Well some features happen more than once! Ellie has two ears, two eyes, four paws, etc. If we know what defines a paw at one place in a sequence it will be just as useful elsewhere should another paw occur!
The Bayesian neural network that I will implement is based off these concepts. If you want to know more about Bayesian neural networks please contact me. Until there is LaTeX support, I can not type set mathematics :P
If you are curious as this might work, I will try to give a conceptual overview. So we have seen how a convolutional neural network (CNN) uses LRFs to abstract features across as sequence. Well humans are made of sequences too! (<------- DNA). Thus the idea is to use A Bayesian neural network (BNN) to learn and predict where exons are. Unlike the ANNs we discussed above, a BNN uses probability measures as its weights, hence we can make a probability distribution about them.
This might make or sense while looking at the image below. A RNA-seq is a genomic sequencing technique. We (academia as a collective) have terabytes (TB) of this data. To put this in perspective, Your genome is about 1.5 gigabyte (GBs), or 2 CDs. That means 1 TB has about 667 genomes, or 1334 CDs! What RNA-seq tells us, is what parts of our genome gets encoded into mRNA (e.g. our exons). Thus using our knowledge of where this mRNA comes from in our genome, we can make an assumption about the distribution of where are exons are, called Priors (B). We can feed this into our BNN (C) to greatly improve its learning and prediction. Its output will give us a new distribution, our posterior beliefs D. We can continue to repeat this process until our network is as great as it can be.
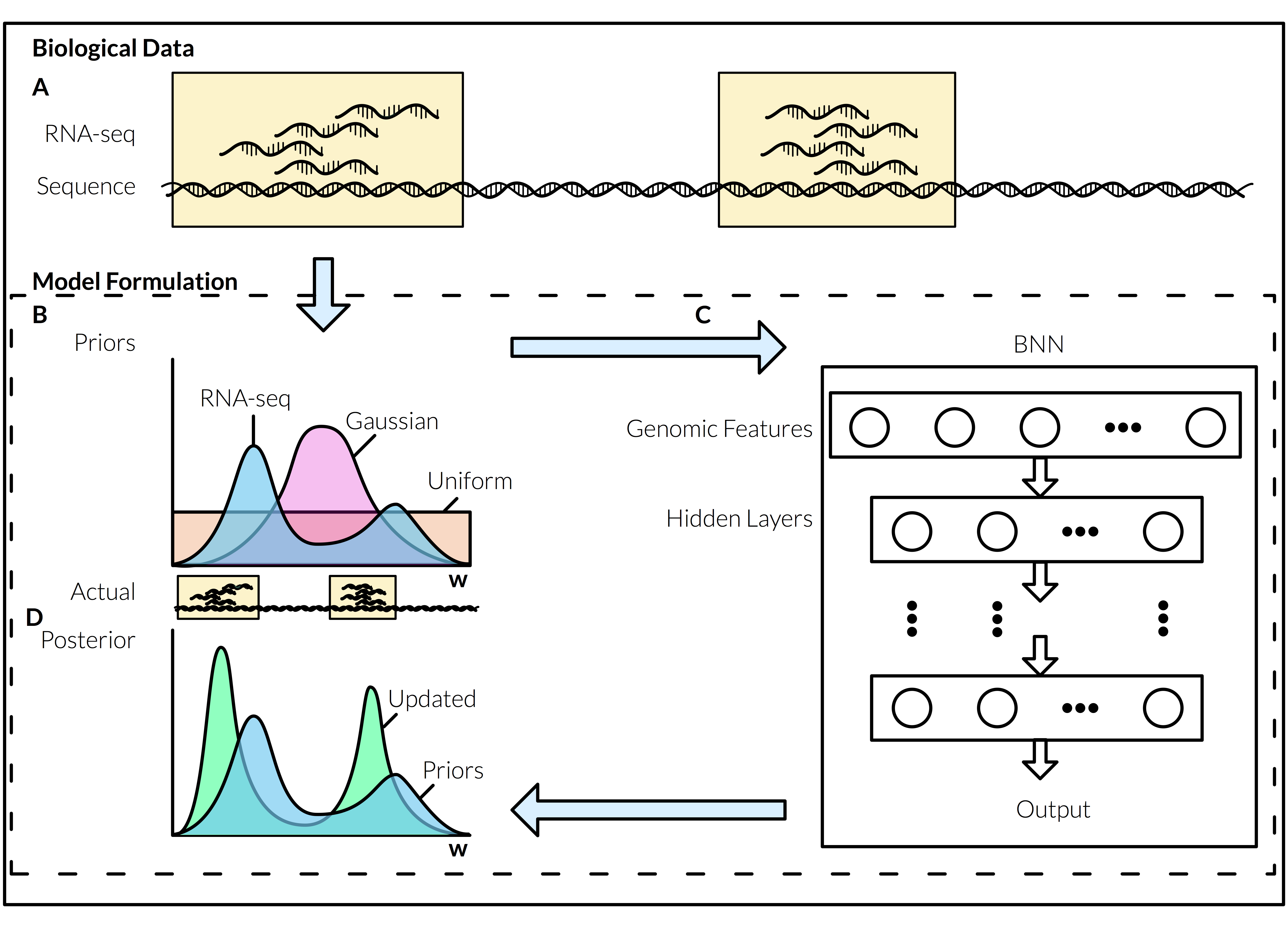
Deep-learning has revolutionized machine-learning. It is what powers Google, and the voice recognition in your smart phone. We have implemented less sophisticated machine-learning techniques with promising results. A very conservative estimate is 1 new exon per gene. That is a lot of new exons (~27,000).
Once we have these exons we can confirm them in vivo. Following all that, this will become a web-app for everyone to use.
Challenges
The main putative challenges are as follows:
- Programming in CUDA (not particularly delightful)
- Finding the optimal architecture for the network
Bayesian neural networks allow for multiple model testing, which resolve the second putative challenge.
Risk factors?
- None. Preliminary work strongly implies it will work. It is just a matter of doing it.
Pre Analysis Plan
Machine learning has a fairly standard way of reporting the findings of models including but not limited to precision and recall, ROC curves, plots of performance verse features, comparison of model verse other machine learning techniques, etc.
The ultimate analysis is in vivo confirmation e.g. take a predicted exon and demonstrate its legitimacy.
Protocols
This project has not yet shared any protocols.