Bioinformatics & Primer Design
To start our exciting journey of figuring out how the Red Devil squid survives extremely low oxygen conditions, we used bioinformatics to create primers for our polymerase chain reaction (PCR) experiment. MicroRNAs bind to specific messenger RNAs to prevent them from turning into protein, which shuts down certain processes in the cell. Our goal is to figure out which microRNAs are more abundant in squids surviving oxygen deprivation compared to the squids that have plenty of oxygen, because this can tell us which processes shut off while they survive “suspended animation”.
So how did we go about doing this with no Red Devil squid genome? Since the Red Devil squid is a mollusk from the class cephalopoda we were able to use a list of known microRNAs from the owl limpet, a type of marine gastropod mollusk. We then compared these microRNA sequences to the nucleotide sequences of other mollusks. This was done by using BLASTn tool to find genomes from each of the bivalve, gastropod, and cephalopod classes so that we could be positive that the sequences would be very similar in the Red Devil squid. 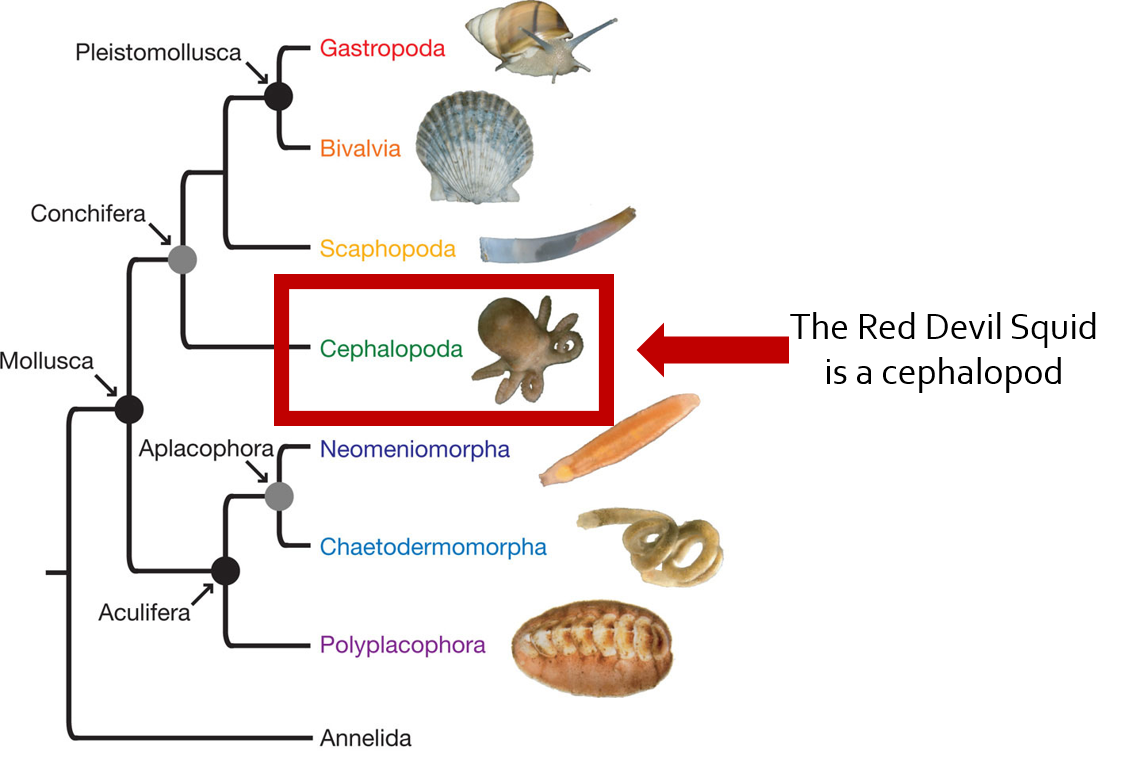
Our bivalve of choice was the pacific oyster, our gastropod was the California sea slug, and our cephalopod was the octopus. Then, we used Clustal Omega to align all of the sequences. The ‘Precursor limpet’ sequence refers to the sequence of the particular microRNA that contains a stem-loop sequence that has yet to be processed into the ‘Mature limpet’ sequence, which refers to the mature microRNA.
Finally, we added a short adapter sequence to the 5’ end (the left end of the sequence) to stabilize our short microRNA primer and allow for amplification. Our final primer looks like this, with our conserved miRNA sequence in bold.
ACACTCCAGCTGGGTATCACAGCCTGCTT
Our primers have been ordered - now we wait.
Stay tuned for our next methods note on RNA extraction and cDNA synthesis!
Samantha, Hanane, and Kenneth
- Published on Aug 31, 2016
- 29 views
- 0 comments
- Print this page
- Back to Methods
