Methods
Summary
Each day, the Red Devil squid is capable of withstanding large fluctuations in the pressure, temperature, and dissolved oxygen content of its environment. Nature has perfected metabolic rate depression in the animal kingdom over millennia and unravelling this blueprint will help further basic biochemical knowledge of gene regulation in animals that are able to adapt to extreme environmental conditions. Using cutting edge techniques we will attempt to elucidate some of the major molecular regulators that give the Red Devil squid its remarkable resilience. To study this we decided to look at the upper-most level of control: epigenetic factors that globally control gene expression by measuring microRNA expression (using qRT-PCR).
MicroRNA discovery:
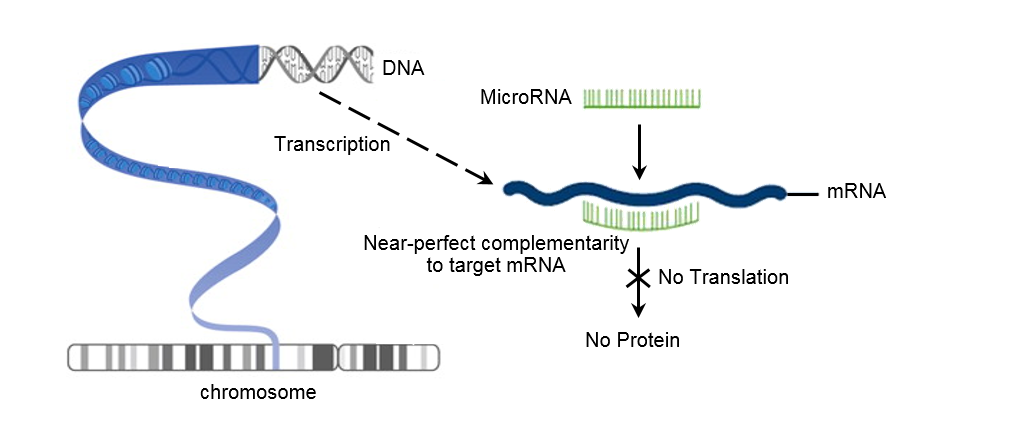
MicroRNAs are a large group of highly-conserved, short, non-coding RNA transcripts (~22 nt). One microRNA can regulate multiple mRNA transcripts, and one mRNA transcript may be subject to regulation by multiple microRNAs. MicroRNAs bind to mRNAs to inhibit protein synthesis by storing the mRNAs in stress granules or by degrading them. To better understand the role that microRNAs play in orchestrating metabolic rate depression in squids, we will take frozen (-80°C) squid tissues (heart, mantle muscle, gut, and potentially brain) and measure microRNA levels in each tissue. We will perform Trizol-based RNA extractions on the active/control and oxygen-deprived squids. The freshly extracted RNA will then undergo polyadenylation, stem-loop adapter ligation, and reverse transcription to convert it into complementary DNA ready for qPCR. Since the Red Devil squid genome has yet to be sequenced we will rely on the genomes of other invertebrates and cephalopods to construct microRNA primers that will work in our invertebrate. We will run RT-qPCR reactions to determine the relative abundance of each microRNA in both control and oxygen-deprived conditions. We will then use bioinformatics target prediction software (DIANA miRPath) to determine which pathways are being actively regulated by microRNAs during the oxygen eprived state of suspended animation.
Protocols
Browse the protocols that are part of the experimental methods.