About This Project
Anopheles arabiensis is a major malaria mosquito in Africa. Some feed on humans, others on cows. Cow-feeders do not contribute to malaria transmission. We've found that a mosquito's genes can determine whether it prefers cows or humans - we're hoping to find the specific genes that promote a human appetite. Manipulating these genes could alter the number of mosquitoes biting people, reducing malaria transmission.Ask the Scientists
Join The DiscussionWhat is the context of this research?
Malaria infects over 200 million people annually,
killing 650,000, mainly children under 5 and pregnant women. Africa bears the heaviest burden; over 90% of malaria deaths occur there. It is caused by a blood parasite transmitted by mosquitoes. Anopheles arabiensis is one of the most important malaria mosquitoes in Africa. However, not all individual A. arabiensis transmit malaria. One critical factor is host preference, what the mosquito feeds on. Some prefer humans making them dangerous malaria vectors. Others prefer cows and so are not malaria transmitters (cows are immune to infection). We recently determined that there is a major genetic determinant to host preference in this mosquito. We now wish to identify the specific genes that influence this critically important mosquito trait.
What is the significance of this project?
We know that malaria mosquitoes bite animals in
addition to people, but we do not know what determines this behavior: is it environmental or genetic? The distinction is important. Can we alter the environment in ways that will divert mosquitoes from people by increasing animal feeding? Alternatively, is this behavior more “hard wired”, can we identify “feeding choice” genes and ultimately identify the chemical cues that turn these genes on? We recently conducted research leading us to the hypothesis that feeding preference has a major genetic component. In this project we aim to test this hypothesis and identify specific “feeding preference” gene candidates, thus establishing the foundation for the development of strategies to manipulate mosquito biting behavior.
What are the goals of the project?
Our overall objective is to identify mutations in specific
genes that are associated with man-biting in the African malaria mosquito, Anopheles arabiensis. We wish to pursue this objective through 3 specific goals:
(1) To design an inexpensive, sensitive and rapid assay that will allow us to identify mutations in up to 40 candidate genes from 90 individual mosquitoes simultaneously.
(2 )To apply this assay to a sample of 1250 individual human and animal fed A. arabiensis mosquitoes that we have already collected from Tanzania.
(3) To analyze the output of these assays and identify those specific genes that are strongly associated with human vs animal feeding.
Budget
Our preliminary study of the genetics of host preference in A. arabiensis is based on examining the whole genome sequences of 65 specimens we collected from villages in the Kilombero Valley of Tanzania. Individual mosquitoes that contained fresh blood in their abdomens were placed in tubes of ethanol and returned to the Vector Genetics Lab at UC Davis. DNA was extracted from the blood within each mosquito and the source of that blood was determined by PCR. Mosquito DNA was extracted from the carcasses of each of these samples and next generation DNA sequencing was conducted to generate sequences for the entire genome of each. Roughly half the specimens had fed on humans and the other on animals, mostly cows. A comparison of their genomes revealed two regions that were highly differentiated.
These results strongly suggest that human vs animal feeding has a genetic basis. In order to confirm this hypothesis and to identify the specific genes contained within the two differentiated regions of the genome we must develop a cost-effective method to identify specific gene mutations in the form of single nucleotide polymorphisms (SNPs) and assay large numbers, hundreds, of animal fed and human fed mosquitoes. We have already collected a sample of 1230 bloodfed mosquitoes from Tanzania and have identified what they fed on (roughly 20% human and 80% animal). We now wish to identify a sample of 40 SNPs from genes within the differentiated regions of the genome. We will design a multiplex SNP genotyping assay using a MALDI-TOF based SNP genotyping system that will detect population origin (human vs animal fed) of a single mosquitoes accurately. As a start, funding of $7,800 will allow us to screen our 1230 individual mosquitoes. The data generated from this work will be used to evaluate our original hypothesis, that feeding preference in this mosquito is genetic and will be the first step in identifying specific genes that influence what an individual mosquito will feed on.
Meet the Team
Team Bio
Gregory Lanzaro (pictured top left) is Professor and head of the VectorGenetics Laboratory (VGL), UC Davis School of Veterinary Medicine. Greg has 20+ years’ experience studying malaria mosquitoes and has conducted research across Africa. His research focuses on the population genomics of insect disease vectors.Yoosook Lee (pictured top right) is a research faculty at the VGL. She is an advocate of open access and is the recipient of the first BioMed Central Open Data Award in 2010. She enjoys getting her hands dirty collecting mosquitoes at field sites in Africa.
Bradley Main (pictured bottom left) is a Postdoctoral Fellow, VGL. His recent work includes a description of the complex genetics of insecticide resistance in African malaria vectors. He spends a lot of his time thinking about mosquito genomes.
Travis Collier (pictured bottom right) is a Programmer at UC Davis who deals with big data. He is one of the main developers of the UC Davis PopI online database.
Gregory Lanzaro
As a child growing up in New York City I spent a lot of time
poring through my uncle’s collection of National Geographic magazine and visiting the American Museum of Natural History. I was especially fascinated by the museum’s insect collection; by the seemingly limitless variety of shapes and colors of these small creatures. I thought how great it would be to travel to remote places and study insects.
I pursued studies leading to a degree in Entomology with a focus in insect genetics. I also developed an interest in parasitology along the way which led me to the field of medical entomology. Insects transmit some of the most dangerous diseases that inflict man, especially in underdeveloped tropical countries.
Today I am lucky enough to have the opportunity to pursue all of these interests as the Head of the Vector Genetics Lab at UC Davis. I work with a team of exceptionally bright people devoted to using modern molecular genetics and genomics to expand our understanding of the relationships between insects and the pathogens they transmit to people, with the ultimate goal of developing new methods to control these deadly diseases.
Our
current work is focused on malaria, the most deadly and widespread mosquito transmitted disease of all. We have had the opportunity to conduct field research throughout Africa, including Mali, Cameroon, Guinea-Bissau, Tanzania, Kenya and the beautiful Comoros Islands located near Madagascar. Just last year we initiated a new program focused on malaria in the Amazon Basin of Brazil. I have a great job!
Bradley Main
I have always been fascinated by the diversity of life on earth and how each organism is perfectly adapted to its natural environment. I'd rather go to a zoo than a theme park any day. I feel very privileged to be a part of a team that is uncovering the genetic basis for specific adaptations or phenotypes that will hopefully facilitate tangible impacts on human health in the near future.
Yoosook Lee
Population Geneticist working on malaria mosquitoes
Press and Media
2007 Award for Excellence in Research : Gregory Lanzaro receives Award for Excellence in Research.BioMed Central Open Data Award: Yoosook Lee wins the first Open Data Award with her publication in the Malaria Journal published in 2009 at the BioMed Central's 4th Annual Research Awards.
2010 Entomological Society of America Professional Awards: Yoosook Lee wins the Journal of Medical Entomology Editor's Choice Award at the 58th Annual ESA Meeting (Lee et al. 2009. JME).
Wingbeats 2011 summer edition features a description of research being conducted at the UC Davis Vector Genetics Laboratory.
UC Davis CAES NewsBeat covers our PNAS paper (Lee et al. 2013).
Microsoft Research Connections Blog: Post about the First Open Data Award sponsored by Microsoft.
BioMed Central Press Release: BioMed Central celebrates excellence in open access publishing.
Entomology & Nematology News covers UC Davis Researchers participating in 2014 Bay Area World Malaria Day Symposium.


Additional Information
This is an Anopheles mosquito that has recently fed on blood. This individual was photographed as it was resting on some clothing hanging in a village hut in rural Mali, west Africa.
The abdomen contains undigested blood which we can use to identify what species of mammal the blood came from (using a PCR assay). The head and thorax is used for extracting the DNA used for whole genome sequencing. With today's next-generation sequencing technology we can sequence the entire 273 million base pair genome from the DNA extracted from a single mosquito.
We have recently completed a study that involved using the methods described above to sequence the genomes of 64 individual A. arabiensis mosquitoes that had fed on either human or cow blood. A comparison of the genomes revealed two discrete genomic regions that largely distinguished cow-fed from human-fed individuals.
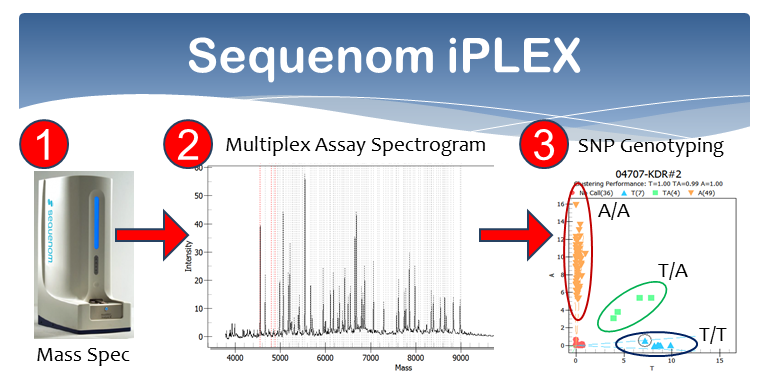
The obvious next step is to (1) verify our results by examining a larger sample of mosquitoes (>1000) and (2) identify specific mutations, in the form of single nucleotide polymorphisms (SNPs), in genes located within the 2 regions of the genome that distinguish human from cow fed mosquitoes. But whole genome sequencing is very expensive at this scale and not necessary now that we have candidate regions. Thus, we propose to develop an inexpensive, but robust multiplex genotyping assay using Sequenom's iPLEX platform (See image below). Identification and characterization of diagnostic SNPs will also provide the foundation for identifying those specific genes and alleles that are influencing host preference in these malaria transmitting mosquitoes.
Project Backers
- 5Backers
- 3%Funded
- $155Total Donations
- $31.00Average Donation



