Methods
Summary
1. A small skin tissue biopsy has been collected from the wing membranes of almost all bats living in Jersey Zoo and Bristol Zoo Gardens since 2011, along with some from carcasses.
2. The skin biopsies will be used to extract DNA in the Molecular Biology Lab at the University of Bristol.
3. Small pieces of DNA will be amplified or duplicated millions of times using a technique called Polymerase Chain Reaction (PCR; fig 1).
4. Each sample will be screened for 12-15 genetic markers to determine the individual identity or fingerprint of each bat.
5. The amplified DNA will be read by a sequencing machine to give us the exact length and value of each DNA fragment (Fig 2).
6. Each bat will be screened three times to have a complete database of all its genetic markers.
7. This information will be used to assemble the pedigree of the population and understand its ancestry and relatedness.
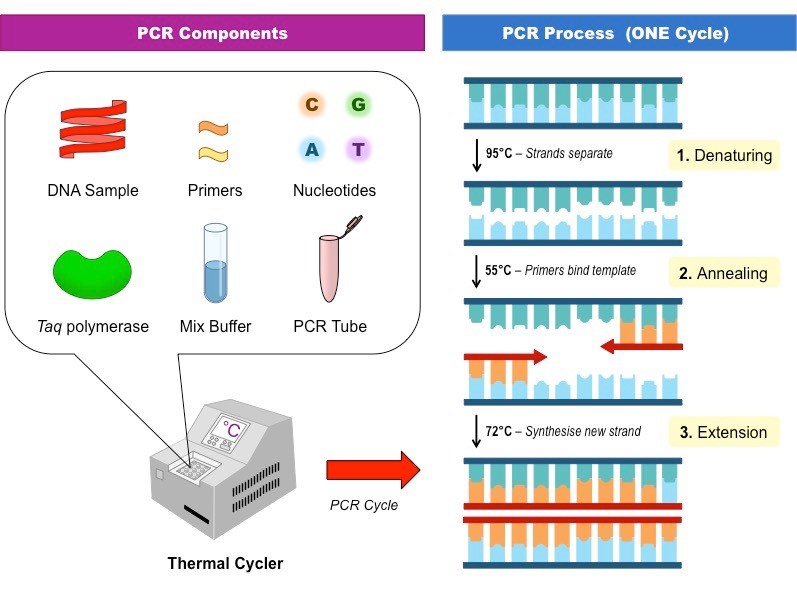
Fig 1. Targeted DNA sequences will be amplified using a Polymerase Chain Reaction (PCR) to create millions of copies that can then be analysed to deduce paternity.
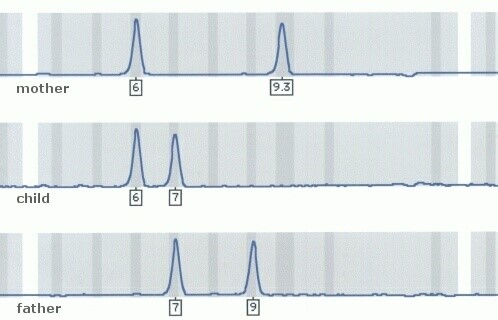
Fig 2. This is an example of the data we will get after the DNA fragments are sequenced. Each peak represents a genetic marker. The numbers represent the number of nucleotides in each fragment and its uniqueness. By looking at the similarity of the peaks, you can deduct that the offspring inherited one genetic marker from each parent.
Challenges
Although DNA is an incredible stable molecule that, under the right conditions, can survive for hundreds of thousands of years, degradation starts the moment an organism dies or when we collect a tissue sample. Cell membranes begin to break down and all sorts of enzymes and chemicals previously safely stored inside the cells are free and start to break down organic compounds, including DNA.
DNA molecules are relatively strong, but their length makes them weak and they are degraded by cutting them into smaller sections. While it is often possible to obtain some DNA from biological materials that have been stored in less than ideal conditions (e.g extinct animals and plants), it is typically difficult to acquire full genomic DNA because sometimes the nucleic acids have begun to degrade and strands of genomic DNA can become more fractured.
For this reason, we need to target several pieces of DNA that will give us enough information about the identity of an organism. We have currently identified 9 very informative strands of DNA and will continue looking for more to create the most accurate ID.
Pre Analysis Plan
Using CERVUS' parentage wizard, paternity can be assigned from sequenced PCR products with a strict confidence level of 95%.
Protocols
This project has not yet shared any protocols.