From Frozen Squid Tissues to microRNAs
Before we can characterize the squid microRNA levels we first had to extract the total RNA from our flash frozen squids. Flash frozen tissues were used to preserve the state of the microRNAs and to ensure that our microRNA measurements reflect what was happening in the animals at the time that they were sacrificed. We weighed approximately 50 mg of active and hypoxic squid brains, muscles, and hearts and then crushed the tissues with a fancy multi-component scientific instrument known as a mortar and pestle.
The frozen tissues were then homogenized (see picture) and RNA was extracted using an organic phenol/chloroform extraction. Tissues are crushed and homogenized to help break the cells open to release the RNA from the cell. Each sample was then centrifuged and separated into three layers- a lower organic layer which contains proteins, an interphase layer which contains DNA, and an upper aqueous layer containing RNA. This top RNA layer was collected and the RNA was precipitated out using isopropanol and much centrifugation. We then measured the absorbance ratios of our RNA to ensure that it was pure. This pure RNA was subsequently separated on an agarose gel to ensure that our RNA was not degraded or contaminated.
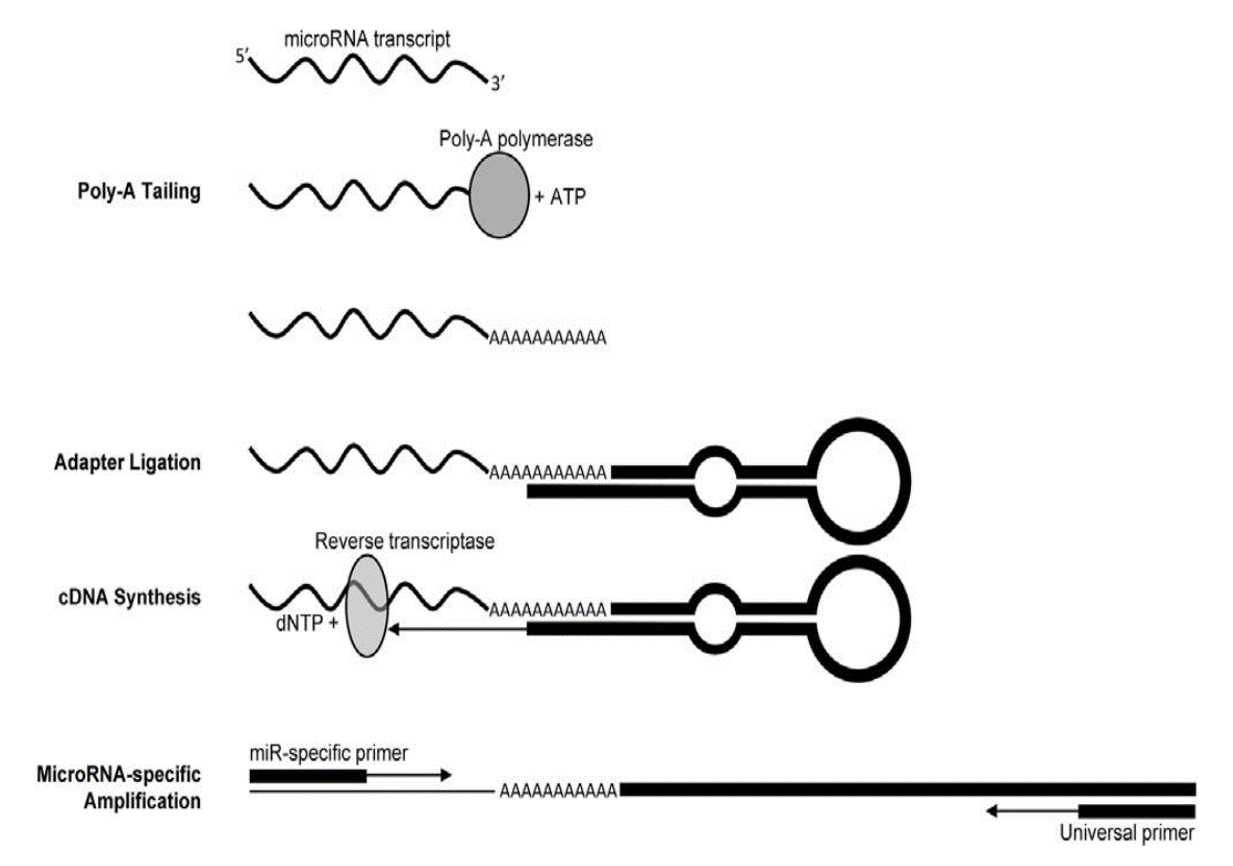
We then prepared 3 µg of this total RNA for qPCR microRNA analysis. To do this, we converted our RNA into complementary DNA, a DNA copy of the RNA, using an enzyme known as reverse transcriptase. But before we converted the RNA to complementary DNA we first added a long tail of adenosine nucleotide bases to our RNA, a process known as polyadenylation that is accomplished using the poly-A polymerase enzyme. We then attached a long synthetic piece of looped DNA, known as a stem-loop adapter, to that poly-A tail. We added this big bulky DNA fragment to our RNA to make our microRNA look bigger, this allows the PCR machine to detect and measure the tiny microRNA fragments. A graphical summary of this process is depicted below (Biggar et al. 2014. Analytical Biochemistry).
Next up? Measure ALL the microRNAs that ever were in squids!
Squid Squad
- Published on Oct 26, 2016
- 10 views
- 0 comments
- Print this page
- Back to Methods