3-2 Bacterial infection and autophagy
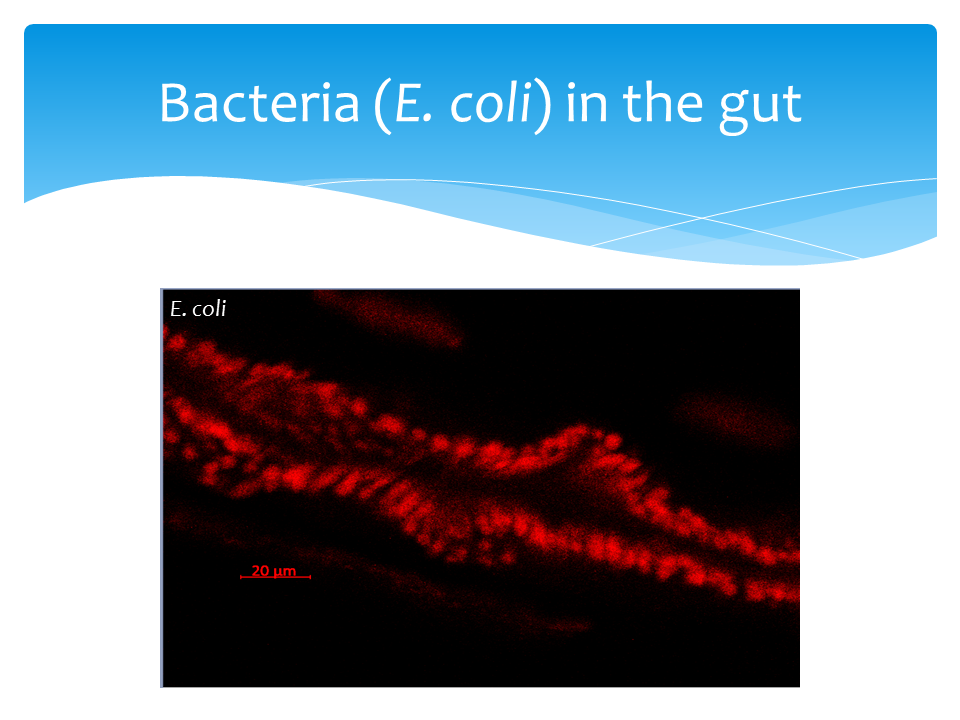
Analysis of bacterial infections presents a complementary inflammatory context to single DSS-induced intestinal injury. We used Escherichia coli (K-12 strain) BioParticle-Alexa 488 conjugate and pHrodo Red E. coli BioParticles Conjugate for Phagocytosis (ThermoFisher, Waltham, MA). Bacteria concentration was titrated and optimized based on intestinal fluorescent intensity. Zebrafish larvae were incubated in 2 μg/μL (6x107 CFU/ml) for one hour. Larvae were rinsed and move to egg water and mortalities were measured and fluorescent at 0, 1, 2, 3 hours.
We used CYTO-ID Autophagy detection kit 2.0 (Enzo) to analyze autophagy with DSS and/or bacterial infection in live zebrafish. Zebrafish were incubated in 1 μL CYTO-ID per 500 μL egg water for one hour with or without E.coli or DSS and wash with 50 mL egg water before imaging. The fluorescent images for quantification were collected upon end of the E. coli treatment with EVOS FL Cell Imaging System. Using ImageJ software, we measured the mean intensity per pixel for the gut and subtract background for individual images. Auto-fluorescence was corrected by comparing to unstained fish gut. The relative expressions (%) were calculated by normalizing to untreated group. Statistical tests were done with Wilcoxon-test in R studio.
- Published on Apr 25, 2018
- 50 views
- 0 comments
- Print this page
- Back to Methods