Methods
Summary
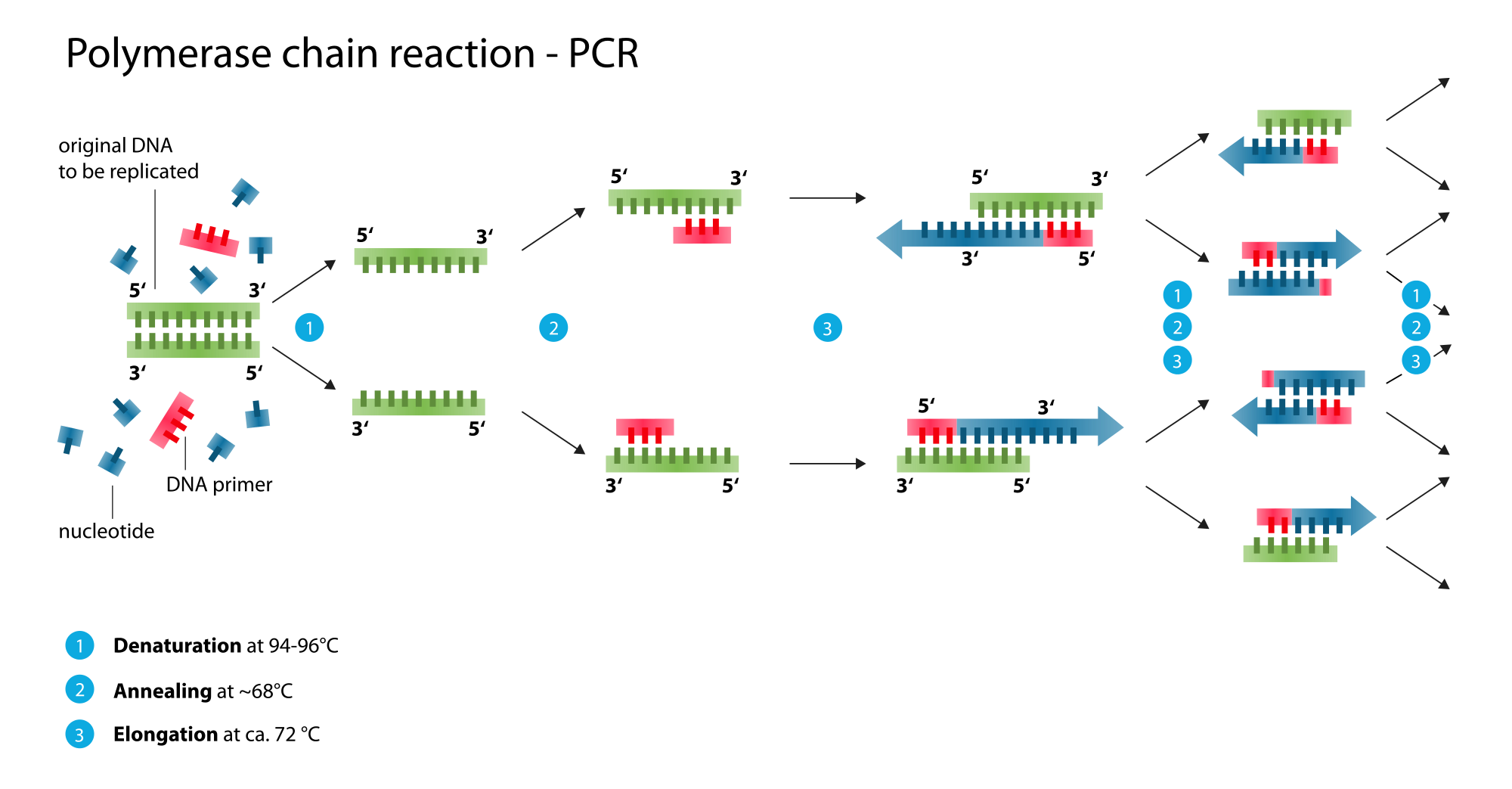
Genetic diversity will be examined using DNA sequencing of the mitochondrial genome (mtDNA) and genotyping of molecular markers called microsatellites in the nuclear genome (nDNA). This will be done by polymerase chain reaction (PCR), a process which amplifies a target fragment of DNA making millions of copies of the DNA of interest.
We can then read the fragment using capillary electrophoresis by either sequencing (mtDNA) or genotyping (nDNA).
DNA isolation, PCR and DNA sequencing and analysis will be completed using standard laboratory technologies in the DNA Technologies Core Laboratory at Texas A&M University in College Station, TX.
Preliminary data for this project can be found in this publication:
Curry CJ, White PA, Derr JN (2015) Mitochondrial Haplotype Diversity in Zambian Lions: Bridging a Gap in the Biogeography of an Iconic Species. PLoS ONE 10(12): e0143827. doi:10.1371/journal.pone.0143827Challenges
Microsatellite primers used for African lions were initially described for the domestic cat but have been used successfully in multiple studies on the African lion. However, using bioinformatics, I have discovered that these primers are not all exact matches which can cause some issues when using DNA from historical specimens, or aDNA. Over time DNA begins to degrade breaking the DNA into shorter fragments which can be more difficult to amplify during PCR. One way to solve this issue is by making sure your primers are an exact match and as close to your target fragment as possible. So, to increase specificity and reliability, I am redesigning the primers to be specific for the African lion. This redesign is being done through both molecular (PCR and sequencing) and bioinformatic (computer analysis) methods.
Pre Analysis Plan
Statistical analysis will consist of the calculation of: percentage of polymorphic loci, number of alleles per locus, observed and expected heterozygosity (HE and HO), and number of unique alleles, deviations from Hardy-Weinberg Equilibrium, the coefficient of differentiation, and nucleotide and haplotype diversity. Population structure will be examined through Bayesian clustering and determination of effective population size.
This study aims to access levels of genetic diversity across time (years) and space (geographical distance). Through analysis of mtDNA and microsatellites we can detect historic changes in population size and diversity, the effects of population fragmentation, and genetic exchange between populations.
Protocols
This project has not yet shared any protocols.