A General Transformation Procedure for C. reinhardtii using the Rescue of a Photosynthetic Knock-out Mutant for Selection
Background
The following protocol is for the transformation of a green alga, C. reinhardtii, using the rescue of a photosynthetic mutant. This method is advantageous over the use of an antibiotic marker since it avoids the controversial issues of releasing foreign antibiotic resistance genes into the wild. The method relies on using an expression vector (pASapI) with a native gene as the selectable marker (Figs. 1-3 below). It is best to use a mutant strain of chlamydomonas that lacks a cell wall (i.e. cw15), but one can also make cells competent using a proteolytic mating enzyme. Instructions on how to culture this enzyme by mating two wild-type algae strains can be found here.
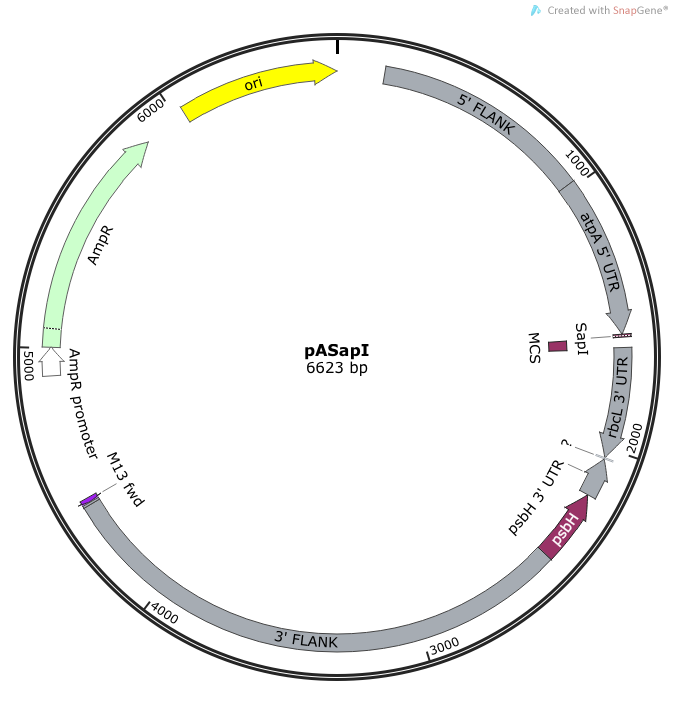 Fig. 1: the pASapI plasmid. Note in this plasmid, the restorative photosynthetic gene, psbH, which allows a transformed algae cell to survive on minimal media.
Fig. 1: the pASapI plasmid. Note in this plasmid, the restorative photosynthetic gene, psbH, which allows a transformed algae cell to survive on minimal media.
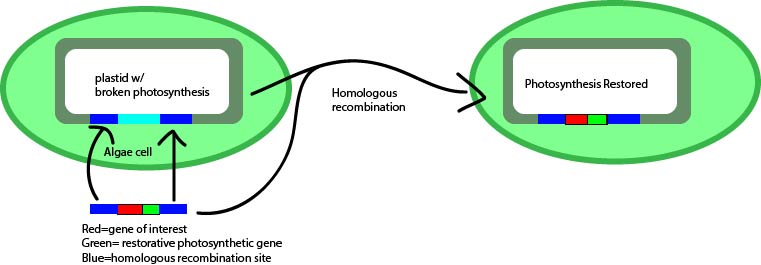 Fig. 2: Illustration showing the insertion of a gene of interest and a restorative photosynthetic gene for the selection process.
Fig. 2: Illustration showing the insertion of a gene of interest and a restorative photosynthetic gene for the selection process.
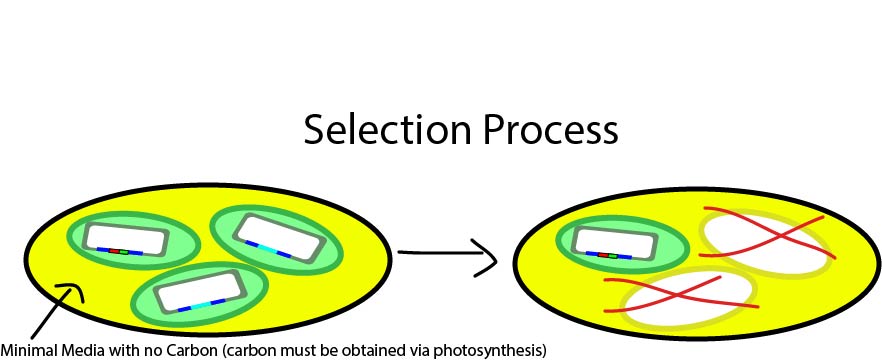 Fig. 3: An illustration of the selection process. Untransformed algae cells do not survive on minimal media, which requires that carbon be fixed from CO2, which can only be done if the photosynthetic gene is restored.
Fig. 3: An illustration of the selection process. Untransformed algae cells do not survive on minimal media, which requires that carbon be fixed from CO2, which can only be done if the photosynthetic gene is restored.
Materials
1. The following strains of the microalgae, Chlamydomonas reinhardtii are required to perform this procedure (purchase links are also provided):
For the production of proteolytic mating enzyme to remove cell wall, gametolysin (search google for preparation instructions):
CC620 (mt+ wildtype, high efficiency mating)
CC621 (mt- wildtype, high efficiency mating)
The photosynthetic mutant
2. The following growth media components are required:
a. Purified laboratory-grade water. Alternatively, but less ideal, distilled water may work as well.
b. If you have the money, pre-made TAP media is good, but for the more economically-conscience, High Salt Medium works just as well, if not better.
c. Bacto agar
d. Purified Water
3. The following DNA plasmid vectors and reagents are required:
a. Your pASapI plasmid with recombinant gene ligated
Procedure
1. Prepare 300mL of HSMA (High Salt Medium + Acetate), as described, in a 1000mL flask, with the addition of 2g/L of sodium acetate for the carbon source. This will be your source stock of photosynthetic mutants.
2. In a sterile environment, inoculate the mixture with a small swab of psbH mutant cells
3. Grow starter liquid cultures overnight.
4. Transfer 4mL of starter culture into 400mL of HSMA and continue to grow cells for a week, or to a density of 1*10^6 cells/mL).
5. Remove liquid and resuspend cells in 3mL of HSM (no acetate) into a medium-sized beaker.
6. In your experimental design, include a negative control. Transfer cell suspension into two separate flasks.
7. Add 5ug of plasmid DNA to your HSM flask suspension
8. Add 0.3 grams of sterile glass beads and agitate using a vortexer at max speed for 15 seconds.
7. Prepare HSM+agar plates (for selection), not neglecting your positive (HSMA) and negative (HSM, no plasmid) controls
8. Agitate cells for 15 seconds using a vortexer at maximum speed and spread them on the selection plates.
9. Prepare HSM plates (No acetate) with 2g/L bacto agar. (These plates, which rely on phototrophic growth, will select for restorative pASAPI plasmid.
10. Incubate plates at 25 degrees celcius in dim light overnight, and then transfer to bright light and incubate for two-three weeks, until colonies appear.
11. Verify transformation results using polymerase chain reaction (PCR)
Acknowledgements
More information can found in this procedure by referring to the excellent paper published by Economou, C, et. al.
Preparation of Gamete Autolysin. Chlamydomonas Resource Center.
Economou, C., Wannathong, T., Szaub, J., and Purton, S. (2014). A simple, low-cost method for chloroplast transformation of the green alga Chlamydomonas reinhardtii. Methods Mol. Biol. 1132, 401–411.
- Published on Dec 29, 2015
- 346 views
- 0 comments
- Print this page
- Back to Methods